OptiSeq™ NGS Targeted Sequencing Service
End-to-end NGS solution and gene panels powered by XNA technology
No more costly and time-consuming deep sequencing! DiaCarta’s OptiSeq™ Next-Generation sequencing platform removes the cost and time barriers by reducing the sequencing depth by a factor of up to 100X and enabling ultra-sensitive detection of genetic mutations. The platform uses the proprietary XNA technology that only hybridizes the wild-type template. That way, only the mutant template is amplified and sequenced.
500 sequencing reads on the OptiSeq™ platform are equivalent to 50,000 reads without using the platform.
Research-use-only validated panels
OptiSeq™ Pan-Cancer Panel (65 Genes)
DiaCarta offers a sample-to-report OptiSeq™ Targeted Sequencing cancer diagnostic service at its state-of-the-art CLIA certified facility. The liquid biopsy/FFPE sample prep workflow provides clients with fast turnaround time and accurate VAF detection process.
OptiSeq™ Colorectal Cancer NGS Panel (43 Genes)
DiaCarta offers a sample-to-report OptiSeq™ Targeted Sequencing cancer diagnostic service at its state-of-the-art CLIA certified facility. The liquid biopsy/FFPE sample prep workflow provides clients with fast turnaround time and accurate VAF detection process.
Other Panels
OptiSeq™ Lung Cancer Panel (32 Genes)
Including AKT1, ALK, BRAF, CCND1, CDKN2A, CTNNB1, DDR2, EGFR, EIF1AX, ERBB2, ERBB4, FGFR1, FGFR2, FGFR3, GNAS, HRAS, IDH1, IDH2, KIT, KRAS, MAP2K1, MDM2, MET, NRAS, PDGFRA, PI3CA, PTEN, RET, ROS1, STK11, TP53, TSHB
OptiSeq™ Lung Cancer & Colon Cancer Early Detection Panel (6 Genes)
Including CTNNB1, KRAS, NRAS, APC, EGFR and BRAF.
OptiSeq™ BRCA1 and BRCA2 Panel (2 Genes)
Including BRCA1 and BRCA2.
OptiSeq™ NGS Targeted Sequencing Service Supporting Data
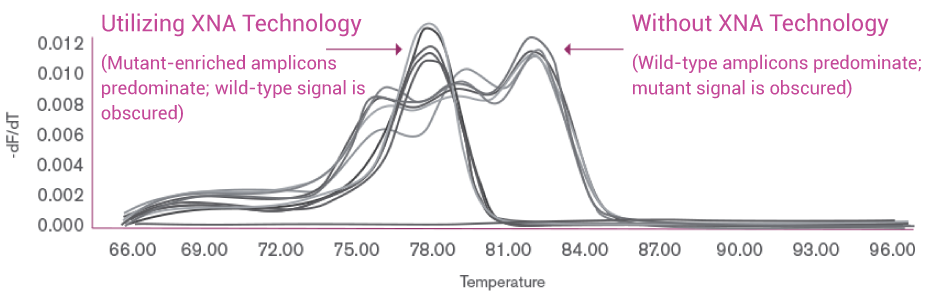
Melting Profiles of PCR Amplicon
OptiSeq™ applies XNA technology to predominantly enrich the KRAS codon 12 mutant allele from a lung tumor sample, resulting in a pure mutant PCR amplicon as indicated by a single peak in the high resolution melting profile (on the left). PCR for conventional NGS without XNA technology, on the other hand, obscures the mutant allelic signal in the profile (on the right).
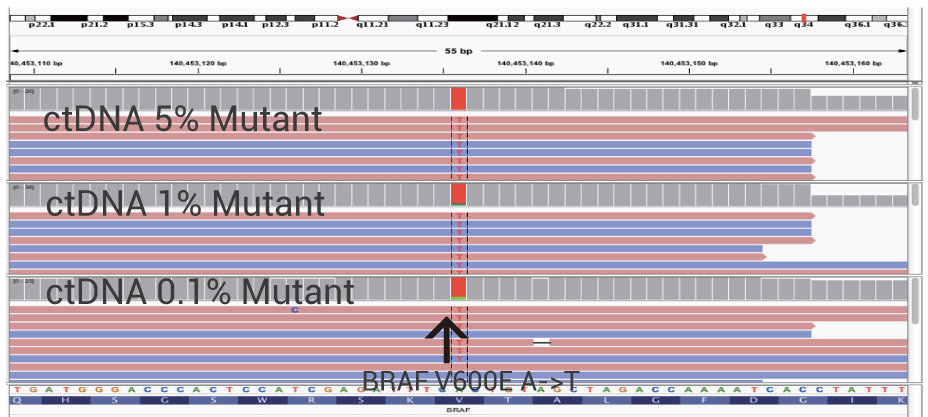
Ultra-Sensitive Detection of BRAF V600 Mutation
Ultra-sensitive detection of rare and actionable mutations is achieved without deep sequencing. OptiSeq™ rapidly, precisely and cost-effectively detects the BRAF V600E mutation from ctDNA in whole blood. The detection levels of 5%, 1% and 0.1% mutant DNA are shown.
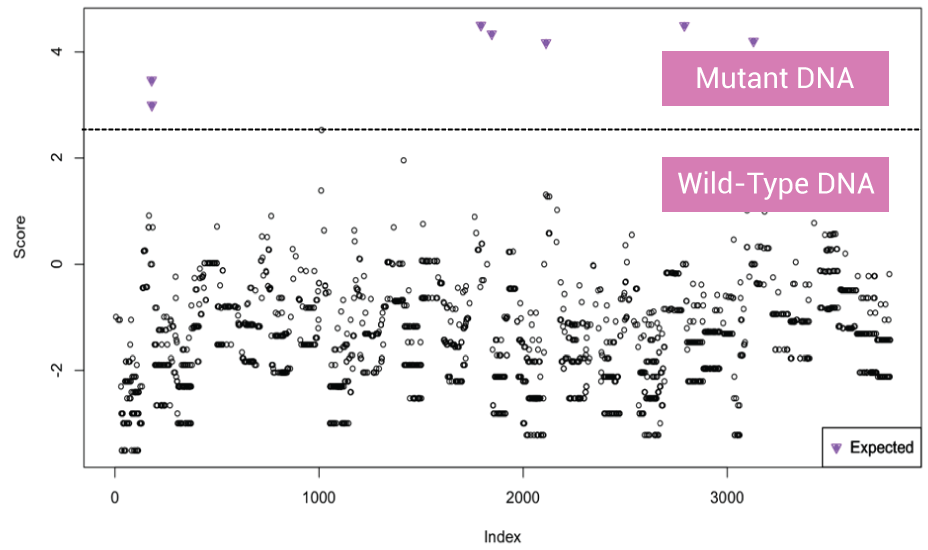
OptiSeq™: Background Noise Reduction
Data coming from circulating tumor DNA mutation sites present high background noise from wild-type DNA (filled and open circle below the dotted line) separated from mutant DNA signal (inverted triangle above dotted line). For mutant DNA, only variant DNA template sequences are sequenced/analyzed. 500 reads OptiSeq™ = 50,000 reads without XNA technology. For wild-type DNA, wild-type alleles are not analyzed, so it reduces time but still gives high-quality read depth.
Advantages of OptiSeq™ NGS Targeted Sequencing Service

Ultra-Sensitive
Ability to detect below 0.1% mutations

Sample Format
Suitable for FFPE tissue (e.g. solid tumor) and blood samples (e.g. ctDNA)
Fast Result
Streamlined workflow with 7-10 days turnaround time from sample to report
Accurate
Fewer errors by enriching mutant allele reads

Reproducible
Robust XNA technology minimizes variability
Cost-Effective
Achieved by reducing 100x read depth
Streamlined Workflow for OptiSeq™ NGS Targeted Sequencing Service

Step 1: Sample prep & QC
Step 2: Target enrichment
Step 3: Library Preparation & QC & Template
Step 4: Sequencing Run
Step 5: Data Analysis
Step 6: Reporting
OptiSeq™ NGS Targeted Sequencing Service Specifications
Sample Types
FFPE, fresh tissues and liquid biopsy
Target Capture
Hybrid capture protocol
Turnaround Time
10 days from sample preparation to reporting
Target Enrichment
OptiSeq™ target amplification
DNA Library
~80 bp insert DNA library
NGS Platform
MiSeq or GeneReader
